Plot the predictor matrix of an imputation model
Arguments
- data
A predictor matrix for
mice, typically generated with mice::make.predictorMatrix or mice::quickpred, or an object of classmice::mids.- vrb
String, vector, or unquoted expression with variable name(s), default is "all".
- method
Character string or vector with imputation methods.
- label
Logical indicating whether predictor matrix values should be displayed.
- square
Logical indicating whether the plot tiles should be squares.
- rotate
Logical indicating whether the variable name labels should be rotated 90 degrees.
Value
An object of class ggplot2::ggplot.
Details
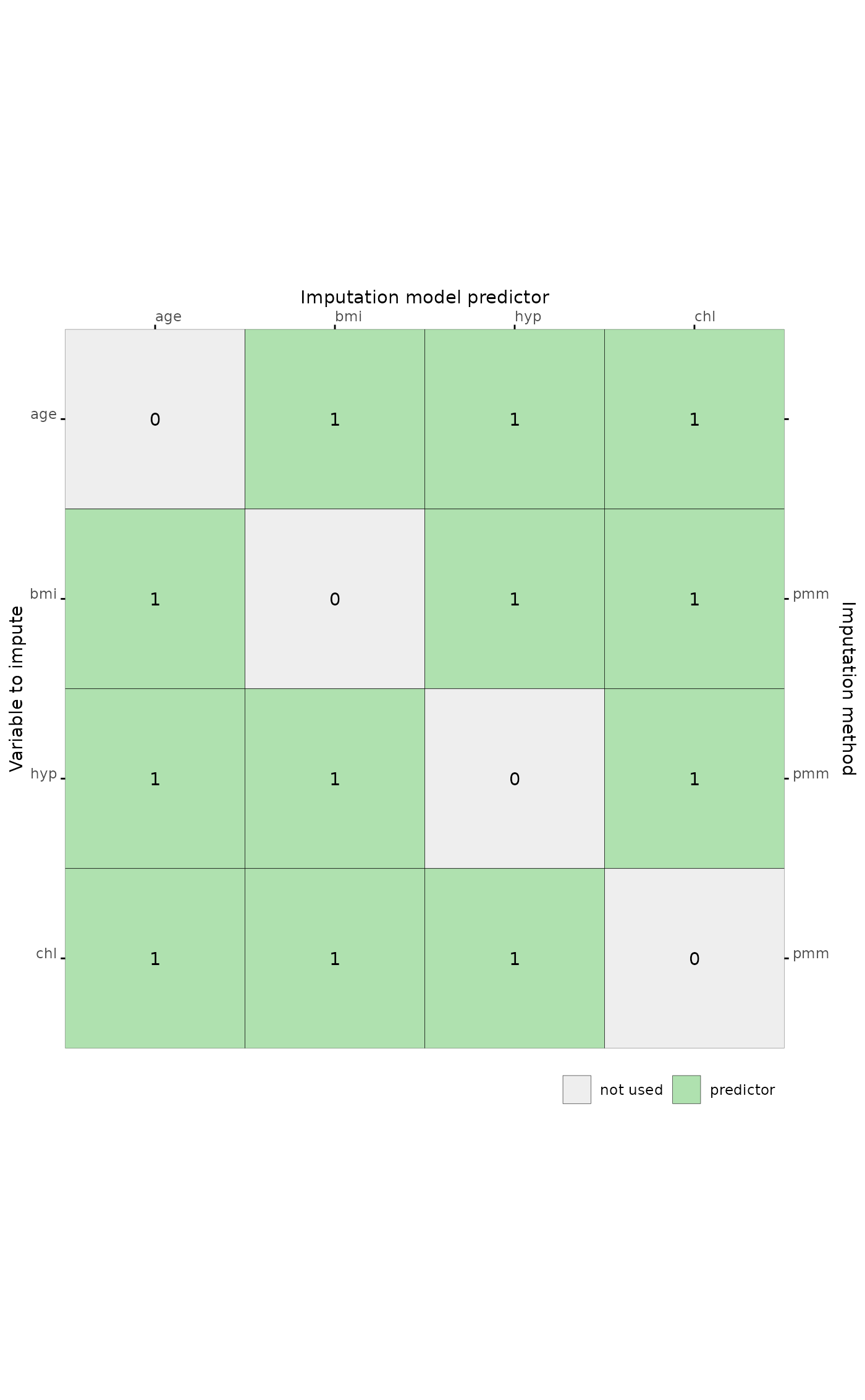
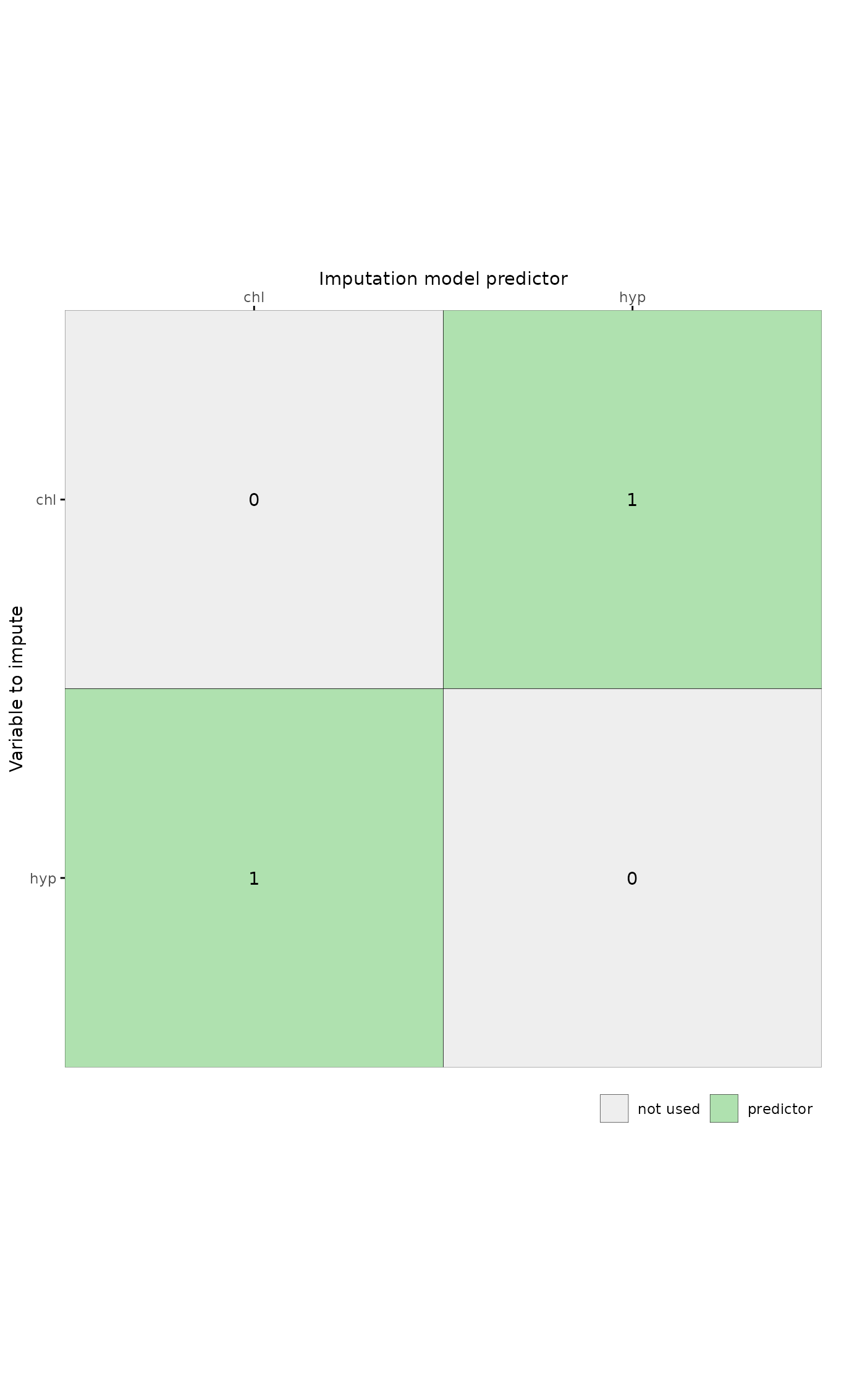
The predictor matrix in mice::mice determines the role an imputation model predictor takes in the imputation model. The rows correspond to incomplete target variables, and the columns to imputation model predictors.
A value of 1 indicates that the column variable is a predictor to impute the target (row) variable. The value 0 means that it is not used as predictor.
Imputation methods for multilevel data use other codes than 0 and 1:
Methods
2l.bin,2l.lmer,2l.norm,2l.pan,2lonly.mean,2lonly.normand2lonly.pmmuse code-2to indicate the class variable;Methods
2l.bin,2l.lmer,2l.normand2l.panuse code2to indicate the random effects;Method
2l.panuses codes3and4to add class means to codes1and2respectively.
References
van Buuren, S. (2018). Flexible imputation of missing data. Chapman and Hall/CRC. stefvanbuuren.name/fimd
Examples
# generate a predictor matrix
pred <- mice::quickpred(mice::nhanes)
# plot predictor matrix for all columns
plot_pred(pred)
#> Ignoring unknown labels:
#> • colour : ""
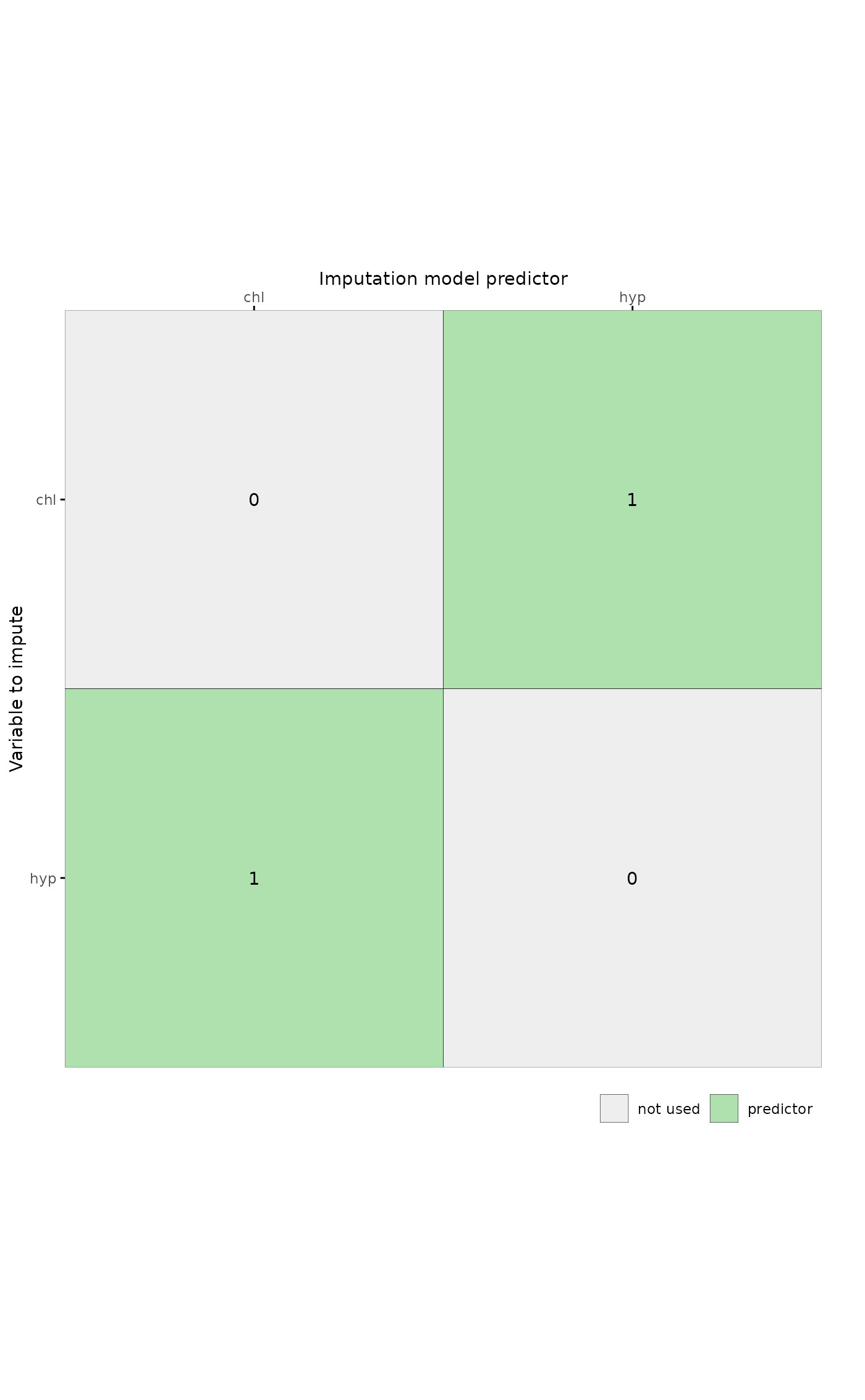
 # plot predictor matrix for specific columns by supplying a character vector
plot_pred(pred, c("chl", "hyp"))
#> Ignoring unknown labels:
#> • colour : ""
# plot predictor matrix for specific columns by supplying a character vector
plot_pred(pred, c("chl", "hyp"))
#> Ignoring unknown labels:
#> • colour : ""
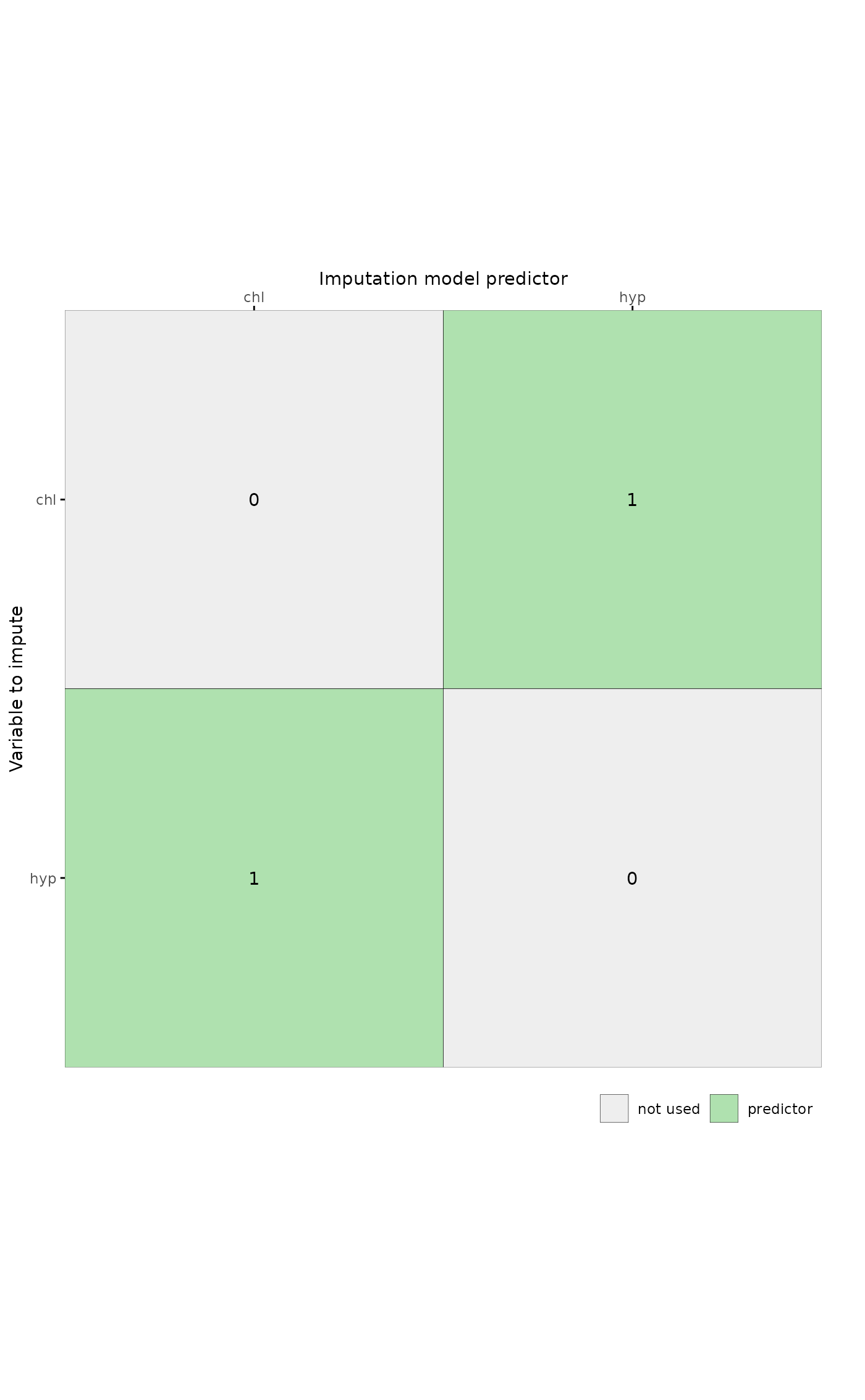
 # plot predictor matrix for specific columns by supplying unquoted variable names
plot_pred(pred, c(chl, hyp))
#> Ignoring unknown labels:
#> • colour : ""
# plot predictor matrix for specific columns by supplying unquoted variable names
plot_pred(pred, c(chl, hyp))
#> Ignoring unknown labels:
#> • colour : ""
 # plot predictor matrix for specific columns by passing an object with variable names
# from the environment, unquoted with `!!`
my_variables <- c("chl", "hyp")
plot_pred(pred, !!my_variables)
#> Ignoring unknown labels:
#> • colour : ""
# plot predictor matrix for specific columns by passing an object with variable names
# from the environment, unquoted with `!!`
my_variables <- c("chl", "hyp")
plot_pred(pred, !!my_variables)
#> Ignoring unknown labels:
#> • colour : ""
 # object with variable names must be unquoted with `!!`
try(plot_pred(pred, my_variables))
#> Error in match_vrb(vrb, row.names(data)) :
#> ✖ The variable name(s) supplied to `vrb` could not be found in `data`.
#> ℹ If you supply an object with variable names from the environment, use `!!` to
#> unqote:
#> `vrb = !!my_variables`
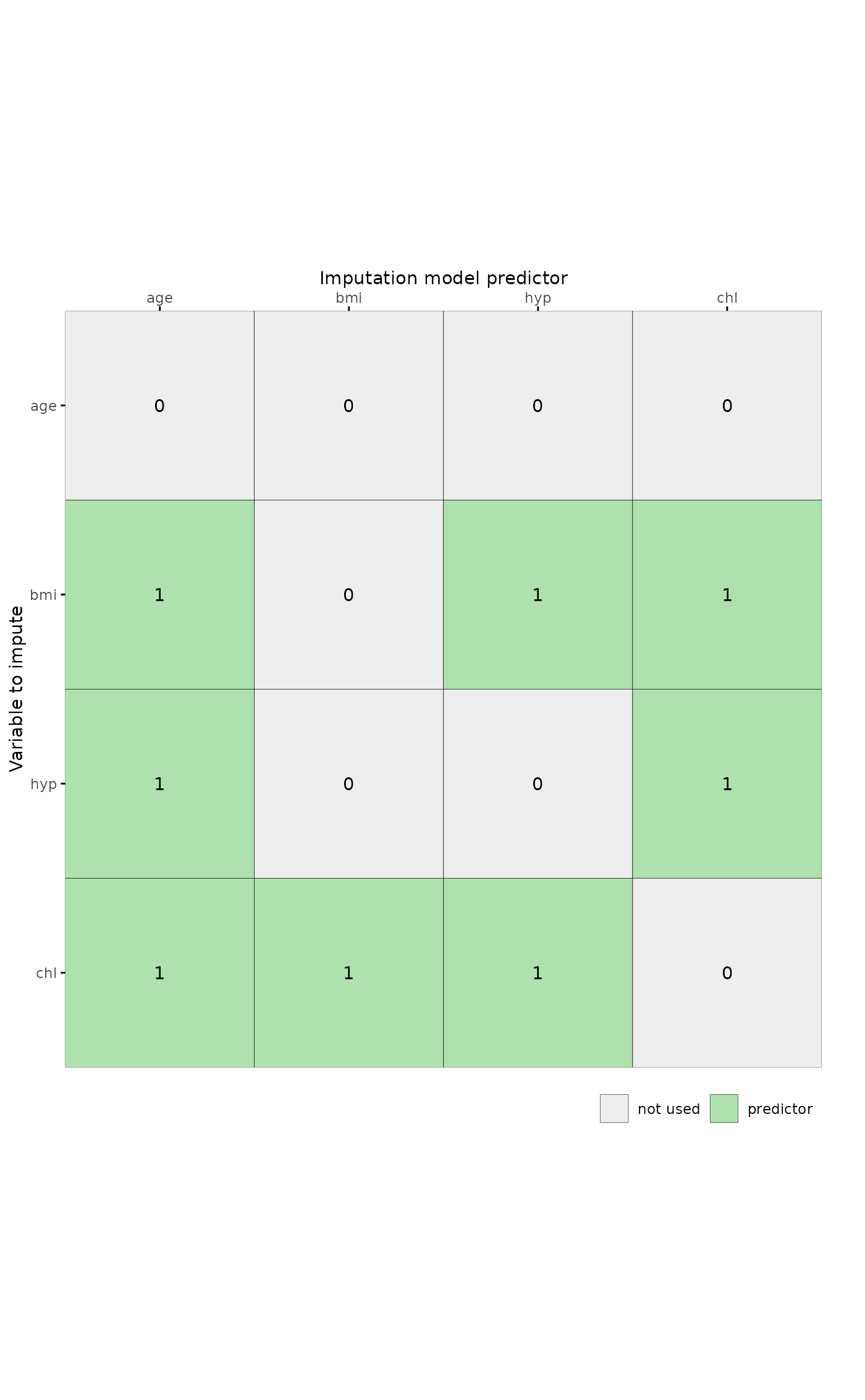
# plot predictor matrix of mids object
imp <- mice::mice(mice::nhanes, print = FALSE)
plot_pred(imp)
#> Ignoring unknown labels:
#> • colour : ""
# object with variable names must be unquoted with `!!`
try(plot_pred(pred, my_variables))
#> Error in match_vrb(vrb, row.names(data)) :
#> ✖ The variable name(s) supplied to `vrb` could not be found in `data`.
#> ℹ If you supply an object with variable names from the environment, use `!!` to
#> unqote:
#> `vrb = !!my_variables`
# plot predictor matrix of mids object
imp <- mice::mice(mice::nhanes, print = FALSE)
plot_pred(imp)
#> Ignoring unknown labels:
#> • colour : ""